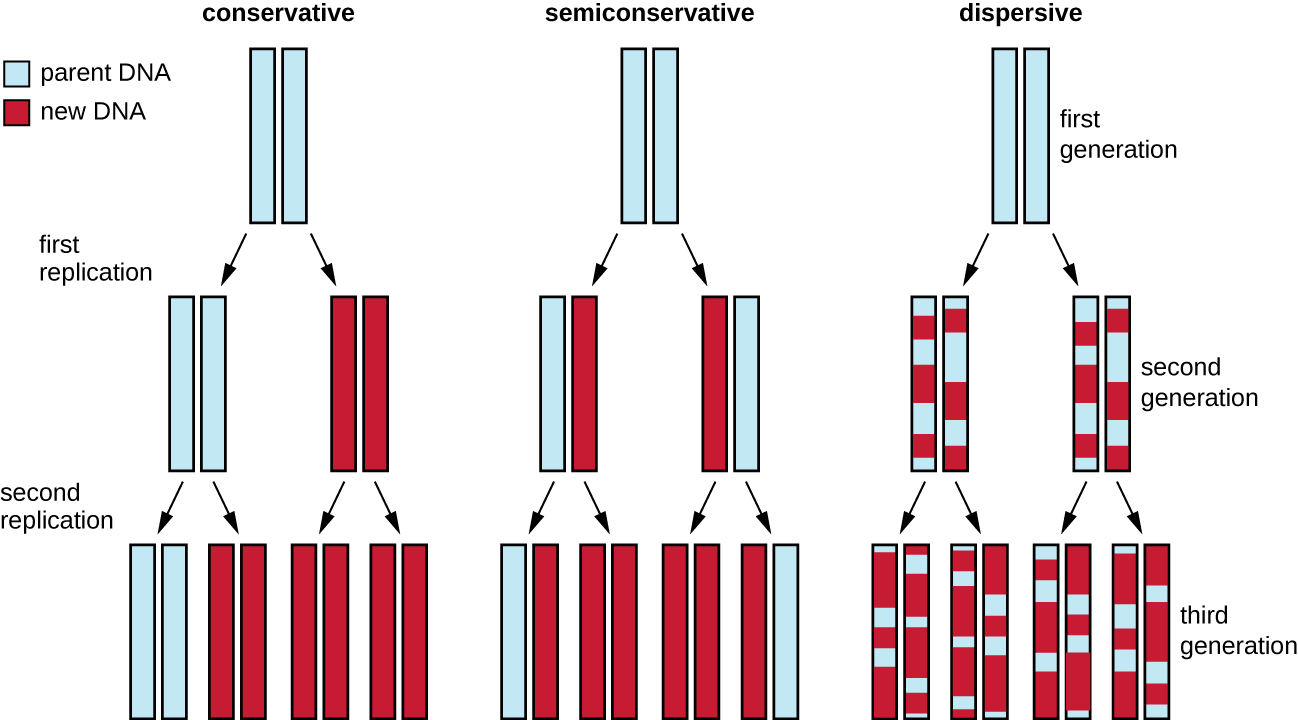
The elucidation of the structure of the double helix by James Watson and Francis Crick in 1953 provided a hint as to how DNA is copied during the process of replication. Separating the strands of the double helix would provide two templates for the synthesis of new complementary strands, but exactly how new DNA molecules were constructed was still unclear. In one model, semiconservative replication, the two strands of the double helix separate during DNA replication, and each strand serves as a template from which the new complementary strand is copied; after replication, each double-stranded DNA includes one parental or “old” strand and one “new” strand. There were two competing models also suggested: conservative and dispersive, which are shown in [link].

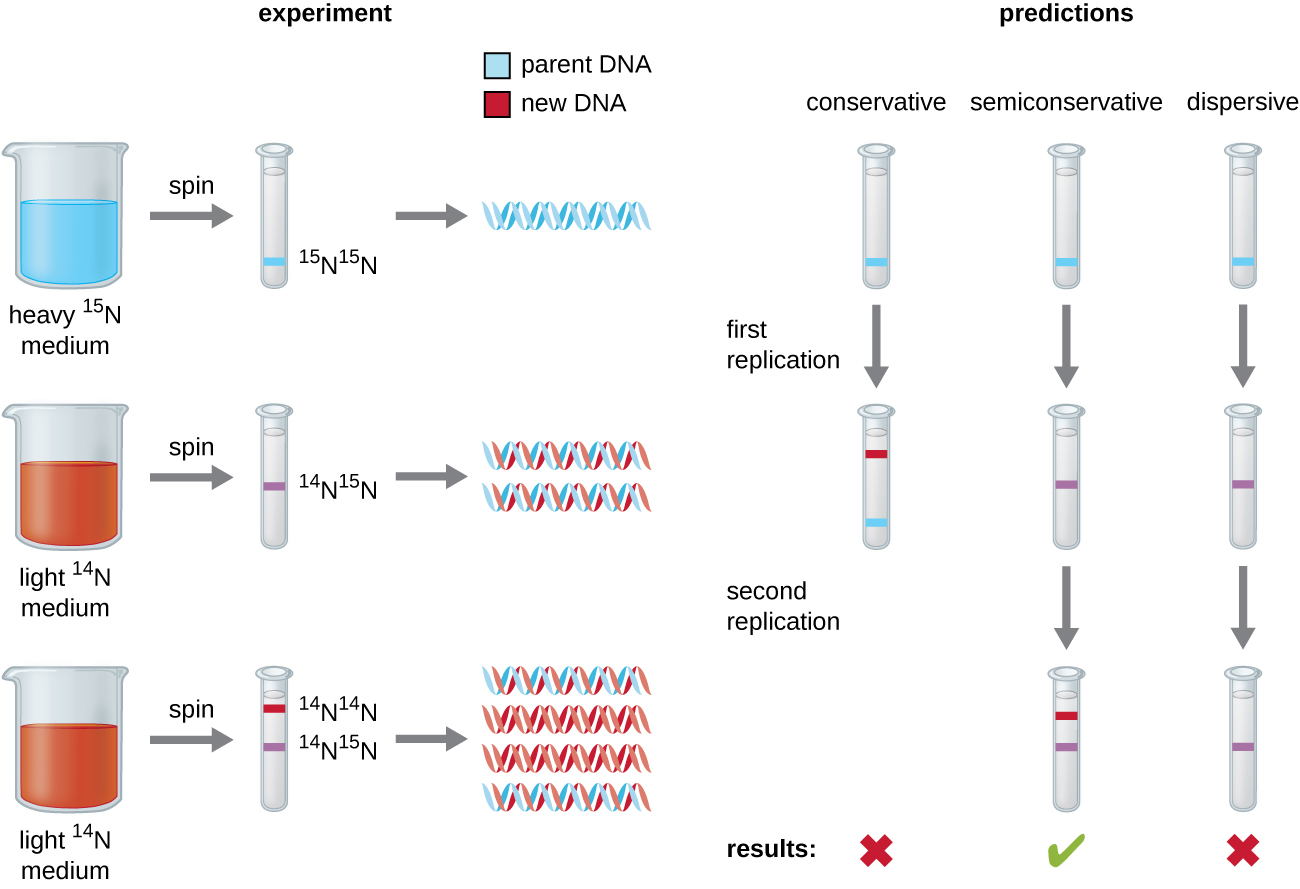
Matthew Meselson (1930–) and Franklin Stahl (1929–) devised an experiment in 1958 to test which of these models correctly represents DNA replication ([link]). They grew E. coli for several generations in a medium containing a “heavy” isotope of nitrogen (15N) that was incorporated into nitrogenous bases and, eventually, into the DNA. This labeled the parental DNA. The E. coli culture was then shifted into a medium containing 14N and allowed to grow for one generation. The cells were harvested and the DNA was isolated. The DNA was separated by ultracentrifugation, during which the DNA formed bands according to its density. DNA grown in 15N would be expected to form a band at a higher density position than that grown in 14N. Meselson and Stahl noted that after one generation of growth in 14N, the single band observed was intermediate in position in between DNA of cells grown exclusively in 15N or 14N. This suggested either a semiconservative or dispersive mode of replication. Some cells were allowed to grow for one more generation in 14N and spun again. The DNA harvested from cells grown for two generations in 14N formed two bands: one DNA band was at the intermediate position between 15N and 14N, and the other corresponded to the band of 14N DNA. These results could only be explained if DNA replicates in a semiconservative manner. Therefore, the other two models were ruled out. As a result of this experiment, we now know that during DNA replication, each of the two strands that make up the double helix serves as a template from which new strands are copied. The new strand will be complementary to the parental or “old” strand. The resulting DNA molecules have the same sequence and are divided equally into the two daughter cells.
DNA replication has been well studied in bacteria primarily because of the small size of the genome and the mutants that are available. E. coli has 4.6 million base pairs (Mbp) in a single circular chromosome and all of it is replicated in approximately 42 minutes, starting from a single origin of replication and proceeding around the circle bidirectionally (i.e., in both directions). This means that approximately 1000 nucleotides are added per second. The process is quite rapid and occurs with few errors.
DNA replication uses a large number of proteins and enzymes ([link]). One of the key players is the enzyme DNA polymerase, also known as DNA pol. In bacteria, three main types of DNA polymerases are known: DNA pol I, DNA pol II, and DNA pol III. It is now known that DNA pol III is the enzyme required for DNA synthesis; DNA pol I and DNA pol II are primarily required for repair. DNA pol III adds deoxyribonucleotides each complementary to a nucleotide on the template strand, one by one to the 3’-OH group of the growing DNA chain. The addition of these nucleotides requires energy. This energy is present in the bonds of three phosphate groups attached to each nucleotide (a triphosphate nucleotide), similar to how energy is stored in the phosphate bonds of adenosine triphosphate (ATP) ([link]). When the bond between the phosphates is broken and diphosphate is released, the energy released allows for the formation of a covalent phosphodiester bond by dehydration synthesis between the incoming nucleotide and the free 3’-OH group on the growing DNA strand.
The initiation of replication occurs at specific nucleotide sequence called the origin of replication, where various proteins bind to begin the replication process. E. coli has a single origin of replication (as do most prokaryotes), called oriC, on its one chromosome. The origin of replication is approximately 245 base pairs long and is rich in adenine-thymine (AT) sequences.
Some of the proteins that bind to the origin of replication are important in making single-stranded regions of DNA accessible for replication. Chromosomal DNA is typically wrapped around histones (in eukaryotes and archaea) or histone-like proteins (in bacteria), and is supercoiled, or extensively wrapped and twisted on itself. This packaging makes the information in the DNA molecule inaccessible. However, enzymes called topoisomerases change the shape and supercoiling of the chromosome. For bacterial DNA replication to begin, the supercoiled chromosome is relaxed by topoisomerase II, also called DNA gyrase. An enzyme called helicase then separates the DNA strands by breaking the hydrogen bonds between the nitrogenous base pairs. Recall that AT sequences have fewer hydrogen bonds and, hence, have weaker interactions than guanine-cytosine (GC) sequences. These enzymes require ATP hydrolysis. As the DNA opens up, Y-shaped structures called replication forks are formed. Two replication forks are formed at the origin of replication, allowing for bidirectional replication and formation of a structure that looks like a bubble when viewed with a transmission electron microscope; as a result, this structure is called a replication bubble. The DNA near each replication fork is coated with single-stranded binding proteins to prevent the single-stranded DNA from rewinding into a double helix.
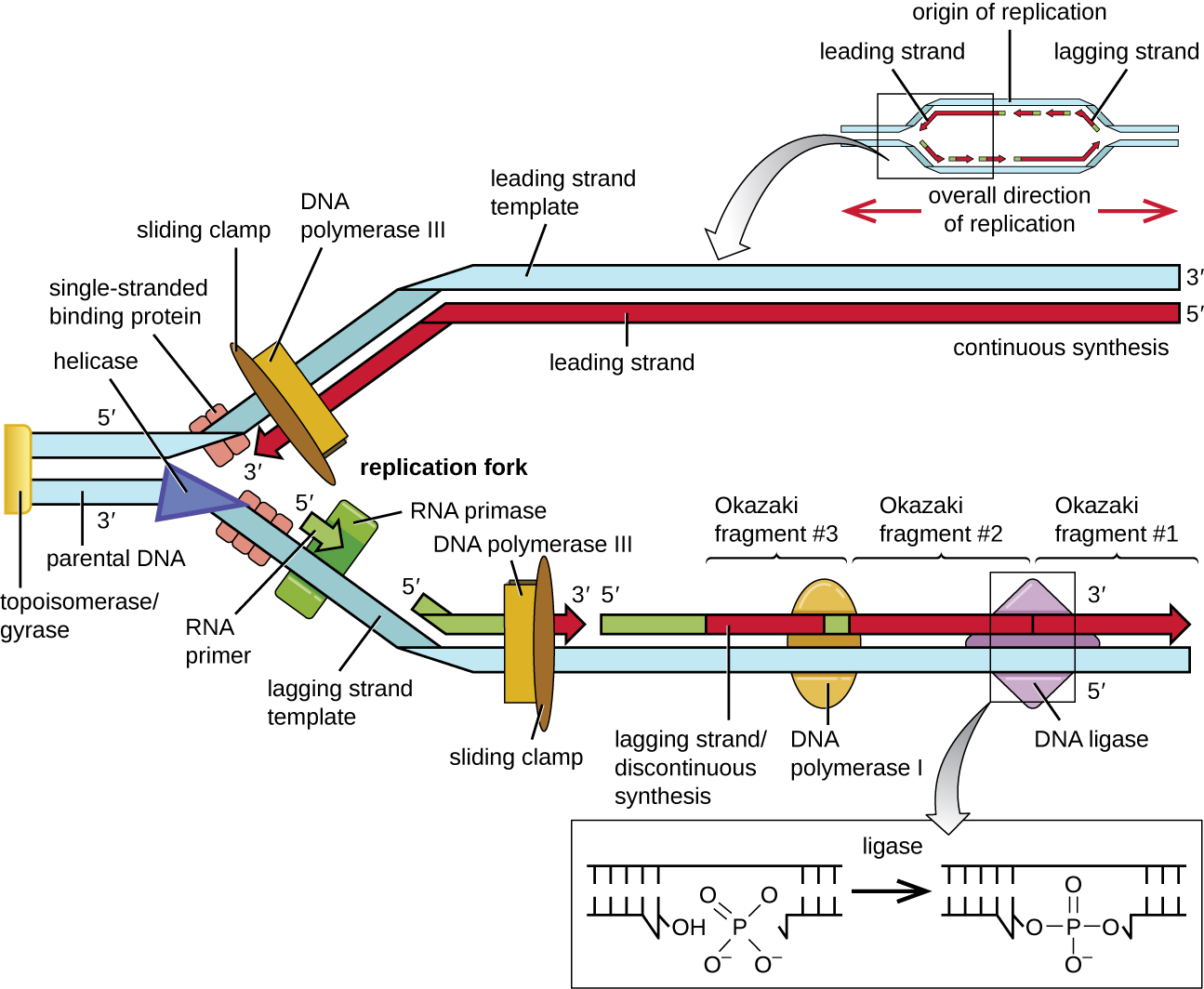
Once single-stranded DNA is accessible at the origin of replication, DNA replication can begin. However, DNA pol III is able to add nucleotides only in the 5’ to 3’ direction (a new DNA strand can be only extended in this direction). This is because DNA polymerase requires a free 3’-OH group to which it can add nucleotides by forming a covalent phosphodiester bond between the 3’-OH end and the 5’ phosphate of the next nucleotide. This also means that it cannot add nucleotides if a free 3’-OH group is not available, which is the case for a single strand of DNA. The problem is solved with the help of an RNA sequence that provides the free 3’-OH end. Because this sequence allows the start of DNA synthesis, it is appropriately called the primer. The primer is five to 10 nucleotides long and complementary to the parental or template DNA. It is synthesized by RNA primase, which is an RNA polymerase. Unlike DNA polymerases, RNA polymerases do not need a free 3’-OH group to synthesize an RNA molecule. Now that the primer provides the free 3’-OH group, DNA polymerase III can now extend this RNA primer, adding DNA nucleotides one by one that are complementary to the template strand ([link]).
During elongation in DNA replication, the addition of nucleotides occurs at its maximal rate of about 1000 nucleotides per second. DNA polymerase III can only extend in the 5’ to 3’ direction, which poses a problem at the replication fork. The DNA double helix is antiparallel; that is, one strand is oriented in the 5’ to 3’ direction and the other is oriented in the 3’ to 5’ direction (see Structure and Function of DNA). During replication, one strand, which is complementary to the 3’ to 5’ parental DNA strand, is synthesized continuously toward the replication fork because polymerase can add nucleotides in this direction. This continuously synthesized strand is known as the leading strand. The other strand, complementary to the 5’ to 3’ parental DNA, grows away from the replication fork, so the polymerase must move back toward the replication fork to begin adding bases to a new primer, again in the direction away from the replication fork. It does so until it bumps into the previously synthesized strand and then it moves back again ([link]). These steps produce small DNA sequence fragments known as Okazaki fragments, each separated by RNA primer. Okazaki fragments are named after the Japanese research team and married couple Reiji and Tsuneko Okazaki, who first discovered them in 1966. The strand with the Okazaki fragments is known as the lagging strand, and its synthesis is said to be discontinuous.
The leading strand can be extended from one primer alone, whereas the lagging strand needs a new primer for each of the short Okazaki fragments. The overall direction of the lagging strand will be 3’ to 5’, and that of the leading strand 5’ to 3’. A protein called the sliding clamp holds the DNA polymerase in place as it continues to add nucleotides. The sliding clamp is a ring-shaped protein that binds to the DNA and holds the polymerase in place. Beyond its role in initiation, topoisomerase also prevents the overwinding of the DNA double helix ahead of the replication fork as the DNA is opening up; it does so by causing temporary nicks in the DNA helix and then resealing it. As synthesis proceeds, the RNA primers are replaced by DNA. The primers are removed by the exonuclease activity of DNA polymerase I, and the gaps are filled in. The nicks that remain between the newly synthesized DNA (that replaced the RNA primer) and the previously synthesized DNA are sealed by the enzyme DNA ligase that catalyzes the formation of covalent phosphodiester linkage between the 3’-OH end of one DNA fragment and the 5’ phosphate end of the other fragment, stabilizing the sugar-phosphate backbone of the DNA molecule.
Once the complete chromosome has been replicated, termination of DNA replication must occur. Although much is known about initiation of replication, less is known about the termination process. Following replication, the resulting complete circular genomes of prokaryotes are concatenated, meaning that the circular DNA chromosomes are interlocked and must be separated from each other. This is accomplished through the activity of bacterial topoisomerase IV, which introduces double-stranded breaks into DNA molecules, allowing them to separate from each other; the enzyme then reseals the circular chromosomes. The resolution of concatemers is an issue unique to prokaryotic DNA replication because of their circular chromosomes. Because both bacterial DNA gyrase and topoisomerase IV are distinct from their eukaryotic counterparts, these enzymes serve as targets for a class of antimicrobial drugs called quinolones.
| The Molecular Machinery Involved in Bacterial DNA Replication | |
|---|---|
| Enzyme or Factor | Function |
| DNA pol I | Exonuclease activity removes RNA primer and replaces it with newly synthesized DNA |
| DNA pol III | Main enzyme that adds nucleotides in the 5’ to 3’ direction |
| Helicase | Opens the DNA helix by breaking hydrogen bonds between the nitrogenous bases |
| Ligase | Seals the gaps between the Okazaki fragments on the lagging strand to create one continuous DNA strand |
| Primase | Synthesizes RNA primers needed to start replication |
| Single-stranded binding proteins | Bind to single-stranded DNA to prevent hydrogen bonding between DNA strands, reforming double-stranded DNA |
| Sliding clamp | Helps hold DNA pol III in place when nucleotides are being added |
| Topoisomerase II (DNA gyrase) | Relaxes supercoiled chromosome to make DNA more accessible for the initiation of replication; helps relieve the stress on DNA when unwinding, by causing breaks and then resealing the DNA |
| Topoisomerase IV | Introduces single-stranded break into concatenated chromosomes to release them from each other, and then reseals the DNA |
Eukaryotic genomes are much more complex and larger than prokaryotic genomes and are typically composed of multiple linear chromosomes ([link]). The human genome, for example, has 3 billion base pairs per haploid set of chromosomes, and 6 billion base pairs are inserted during replication. There are multiple origins of replication on each eukaryotic chromosome ([link]); the human genome has 30,000 to 50,000 origins of replication. The rate of replication is approximately 100 nucleotides per second—10 times slower than prokaryotic replication.
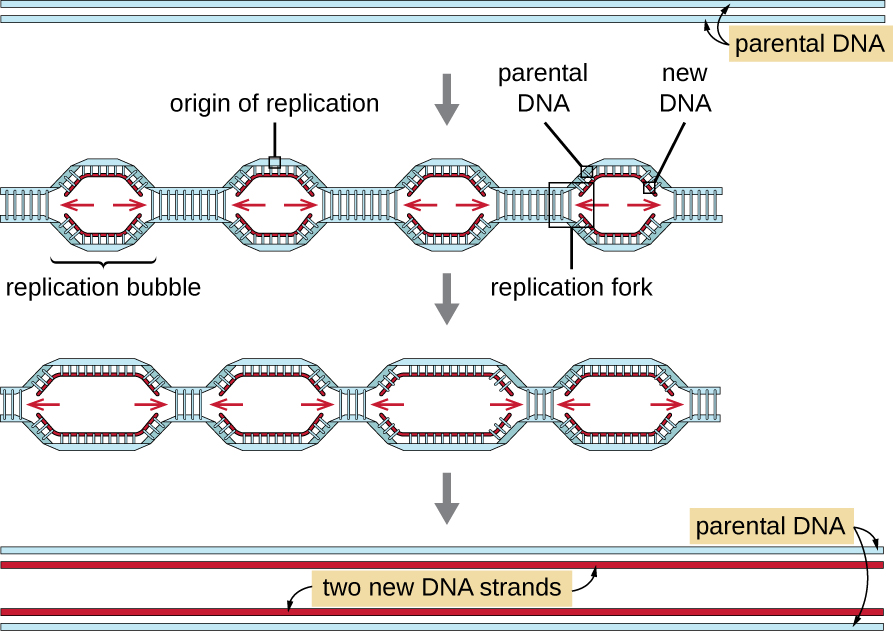
The essential steps of replication in eukaryotes are the same as in prokaryotes. Before replication can start, the DNA has to be made available as a template. Eukaryotic DNA is highly supercoiled and packaged, which is facilitated by many proteins, including histones (see Structure and Function of Cellular Genomes). At the origin of replication, a prereplication complex composed of several proteins, including helicase, forms and recruits other enzymes involved in the initiation of replication, including topoisomerase to relax supercoiling, single-stranded binding protein, RNA primase, and DNA polymerase. Following initiation of replication, in a process similar to that found in prokaryotes, elongation is facilitated by eukaryotic DNA polymerases. The leading strand is continuously synthesized by the eukaryotic polymerase enzyme pol δ, while the lagging strand is synthesized by pol ε. A sliding clamp protein holds the DNA polymerase in place so that it does not fall off the DNA. The enzyme ribonuclease H (RNase H), instead of a DNA polymerase as in bacteria, removes the RNA primer, which is then replaced with DNA nucleotides. The gaps that remain are sealed by DNA ligase.
Because eukaryotic chromosomes are linear, one might expect that their replication would be more straightforward. As in prokaryotes, the eukaryotic DNA polymerase can add nucleotides only in the 5’ to 3’ direction. In the leading strand, synthesis continues until it reaches either the end of the chromosome or another replication fork progressing in the opposite direction. On the lagging strand, DNA is synthesized in short stretches, each of which is initiated by a separate primer. When the replication fork reaches the end of the linear chromosome, there is no place to make a primer for the DNA fragment to be copied at the end of the chromosome. These ends thus remain unpaired and, over time, they may get progressively shorter as cells continue to divide.
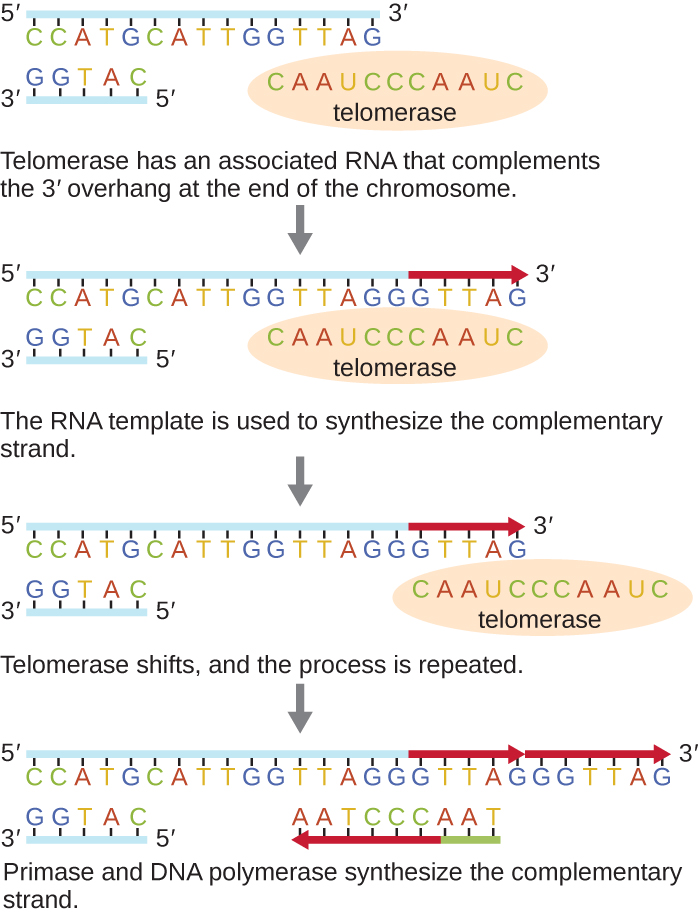
The ends of the linear chromosomes are known as telomeres and consist of noncoding repetitive sequences. The telomeres protect coding sequences from being lost as cells continue to divide. In humans, a six base-pair sequence, TTAGGG, is repeated 100 to 1000 times to form the telomere. The discovery of the enzyme telomerase ([link]) clarified our understanding of how chromosome ends are maintained. Telomerase contains a catalytic part and a built-in RNA template. It attaches to the end of the chromosome, and complementary bases to the RNA template are added on the 3’ end of the DNA strand. Once the 3’ end of the lagging strand template is sufficiently elongated, DNA polymerase can add the nucleotides complementary to the ends of the chromosomes. In this way, the ends of the chromosomes are replicated. In humans, telomerase is typically active in germ cells and adult stem cells; it is not active in adult somatic cells and may be associated with the aging of these cells. Eukaryotic microbes including fungi and protozoans also produce telomerase to maintain chromosomal integrity. For her discovery of telomerase and its action, Elizabeth Blackburn (1948–) received the Nobel Prize for Medicine or Physiology in 2009.
| Comparison of Bacterial and Eukaryotic Replication | ||
|---|---|---|
| Property | Bacteria | Eukaryotes |
| Genome structure | Single circular chromosome | Multiple linear chromosomes |
| Number of origins per chromosome | Single | Multiple |
| Rate of replication | 1000 nucleotides per second | 100 nucleotides per second |
| Telomerase | Not present | Present |
| RNA primer removal | DNA pol I | RNase H |
| Strand elongation | DNA pol III | pol δ, pol ε |
 This animation illustrates the process of DNA replication.
This animation illustrates the process of DNA replication.
To copy their nucleic acids, plasmids and viruses frequently use variations on the pattern of DNA replication described for prokaryote genomes. For more information on the wide range of viral replication strategies, see The Viral Life Cycle.
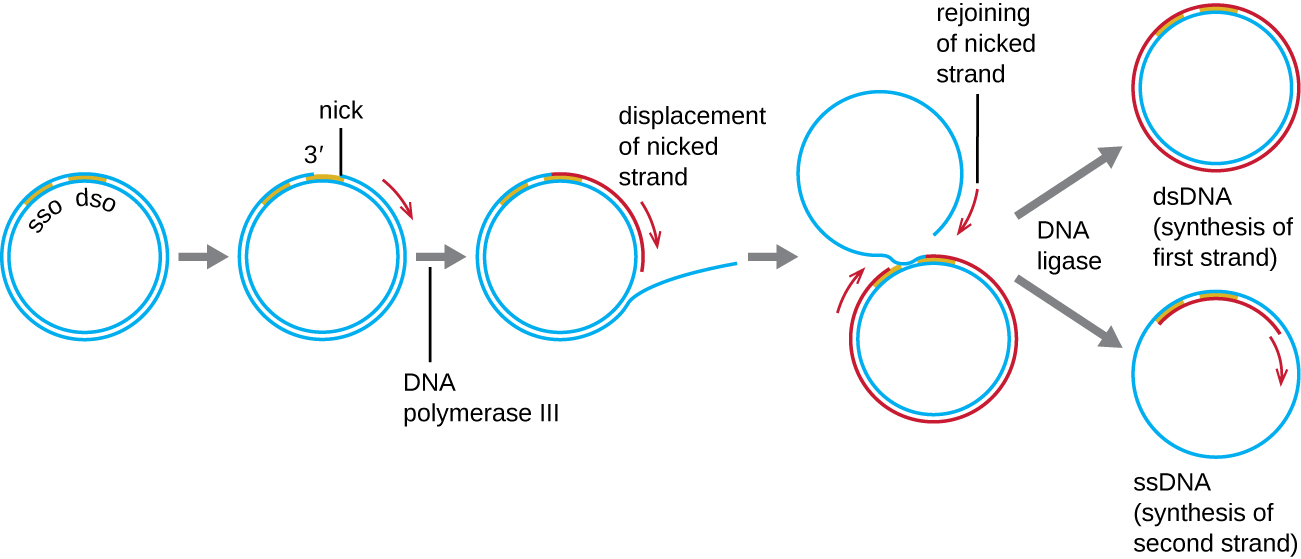
Whereas many bacterial plasmids (see Unique Characteristics of Prokaryotic Cells) replicate by a process similar to that used to copy the bacterial chromosome, other plasmids, several bacteriophages, and some viruses of eukaryotes use rolling circle replication ([link]). The circular nature of plasmids and the circularization of some viral genomes on infection make this possible. Rolling circle replication begins with the enzymatic nicking of one strand of the double-stranded circular molecule at the double-stranded origin (dso) site. In bacteria, DNA polymerase III binds to the 3’-OH group of the nicked strand and begins to unidirectionally replicate the DNA using the un-nicked strand as a template, displacing the nicked strand as it does so. Completion of DNA replication at the site of the original nick results in full displacement of the nicked strand, which may then recircularize into a single-stranded DNA molecule. RNA primase then synthesizes a primer to initiate DNA replication at the single-stranded origin (sso) site of the single-stranded DNA (ssDNA) molecule, resulting in a double-stranded DNA (dsDNA) molecule identical to the other circular DNA molecule.
Which of the following is the enzyme that replaces the RNA nucleotides in a primer with DNA nucleotides?
B
Which of the following is not involved in the initiation of replication?
A
Which of the following enzymes involved in DNA replication is unique to eukaryotes?
D
Which of the following would be synthesized using 5′-CAGTTCGGA-3′ as a template?
C
The enzyme responsible for relaxing supercoiled DNA to allow for the initiation of replication is called ________.
DNA gyrase or topoisomerase II
Unidirectional replication of a circular DNA molecule like a plasmid that involves nicking one DNA strand and displacing it while synthesizing a new strand is called ________.
rolling circle replication
More primers are used in lagging strand synthesis than in leading strand synthesis.
True
Why is primase required for DNA replication?
What is the role of single-stranded binding protein in DNA replication?
Below is a DNA sequence. Envision that this is a section of a DNA molecule that has separated in preparation for replication, so you are only seeing one DNA strand. Construct the complementary DNA sequence (indicating 5’ and 3’ ends).
DNA sequence: 3’-T A C T G A C T G A C G A T C-5’
If deoxyribonucleotides that lack the 3’-OH groups are added during the replication process, what do you expect will occur?

You can also download for free at http://cnx.org/contents/e42bd376-624b-4c0f-972f-e0c57998e765@5.3
Attribution: