By the end of this section, you will be able to:
Prokaryotes and eukaryotes perform fundamentally the same process of transcription, with a few key differences. The most important difference between prokaryotes and eukaryotes is the latter’s membrane-bound nucleus and organelles. With the genes bound in a nucleus, the eukaryotic cell must be able to transport its mRNA to the cytoplasm and must protect its mRNA from degrading before it is translated. Eukaryotes also employ three different polymerases that each transcribe a different subset of genes. Eukaryotic mRNAs are usually monogenic, meaning that they specify a single protein.
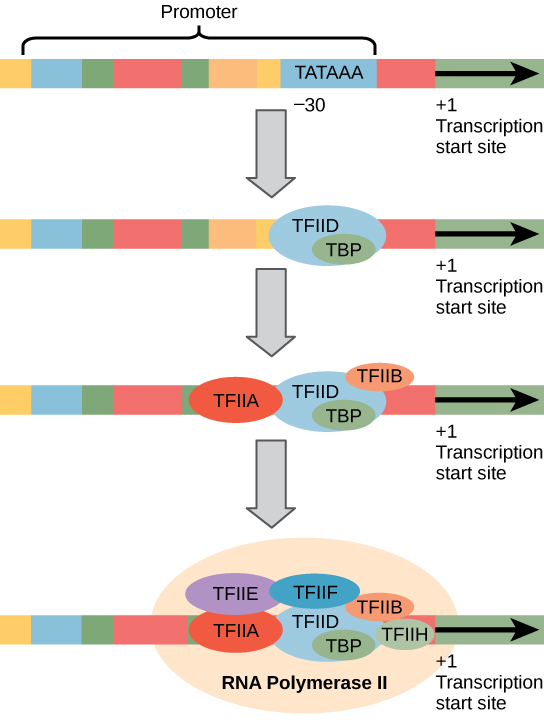
Unlike the prokaryotic polymerase that can bind to a DNA template on its own, eukaryotes require several other proteins, called transcription factors, to first bind to the promoter region and then help recruit the appropriate polymerase.
The features of eukaryotic mRNA synthesis are markedly more complex those of prokaryotes. Instead of a single polymerase comprising five subunits, the eukaryotes have three polymerases that are each made up of 10 subunits or more. Each eukaryotic polymerase also requires a distinct set of transcription factors to bring it to the DNA template.
RNA polymerase I is located in the nucleolus, a specialized nuclear substructure in which ribosomal RNA (rRNA) is transcribed, processed, and assembled into ribosomes ([link]). The rRNA molecules are considered structural RNAs because they have a cellular role but are not translated into protein. The rRNAs are components of the ribosome and are essential to the process of translation. RNA polymerase I synthesizes all of the rRNAs except for the 5S rRNA molecule. The “S” designation applies to “Svedberg” units, a nonadditive value that characterizes the speed at which a particle sediments during centrifugation.
| Locations, Products, and Sensitivities of the Three Eukaryotic RNA Polymerases | |||
|---|---|---|---|
| RNA Polymerase | Cellular Compartment | Product of Transcription | α-Amanitin Sensitivity |
| I | Nucleolus | All rRNAs except 5S rRNA | Insensitive |
| II | Nucleus | All protein-coding nuclear pre-mRNAs | Extremely sensitive |
| III | Nucleus | 5S rRNA, tRNAs, and small nuclear RNAs | Moderately sensitive |
RNA polymerase II is located in the nucleus and synthesizes all protein-coding nuclear pre-mRNAs. Eukaryotic pre-mRNAs undergo extensive processing after transcription but before translation. For clarity, this module’s discussion of transcription and translation in eukaryotes will use the term “mRNAs” to describe only the mature, processed molecules that are ready to be translated. RNA polymerase II is responsible for transcribing the overwhelming majority of eukaryotic genes.
RNA polymerase III is also located in the nucleus. This polymerase transcribes a variety of structural RNAs that includes the 5S pre-rRNA, transfer pre-RNAs (pre-tRNAs), and small nuclear pre-RNAs. The tRNAs have a critical role in translation; they serve as the adaptor molecules between the mRNA template and the growing polypeptide chain. Small nuclear RNAs have a variety of functions, including “splicing” pre-mRNAs and regulating transcription factors.
A scientist characterizing a new gene can determine which polymerase transcribes it by testing whether the gene is expressed in the presence of a particular mushroom poison, α-amanitin ([link]). Interestingly, α-amanitin produced by Amanita phalloides, the Death Cap mushroom, affects the three polymerases very differently. RNA polymerase I is completely insensitive to α-amanitin, meaning that the polymerase can transcribe DNA in vitro in the presence of this poison. In contrast, RNA polymerase II is extremely sensitive to α-amanitin, and RNA polymerase III is moderately sensitive. Knowing the transcribing polymerase can clue a researcher into the general function of the gene being studied. Because RNA polymerase II transcribes the vast majority of genes, we will focus on this polymerase in our subsequent discussions about eukaryotic transcription factors and promoters.
Eukaryotic promoters are much larger and more complex than prokaryotic promoters, but both have a TATA box. For example, in the mouse thymidine kinase gene, the TATA box is located at approximately -30 relative to the initiation (+1) site ([link]). For this gene, the exact TATA box sequence is TATAAAA, as read in the 5' to 3' direction on the nontemplate strand. This sequence is not identical to the E. coli TATA box, but it conserves the A–T rich element. The thermostability of A–T bonds is low and this helps the DNA template to locally unwind in preparation for transcription.

A scientist splices a eukaryotic promoter in front of a bacterial gene and inserts the gene in a bacterial chromosome. Would you expect the bacteria to transcribe the gene?
The mouse genome includes one gene and two pseudogenes for cytoplasmic thymidine kinase. Pseudogenes are genes that have lost their protein-coding ability or are no longer expressed by the cell. These pseudogenes are copied from mRNA and incorporated into the chromosome. For example, the mouse thymidine kinase promoter also has a conserved CAAT box (GGCCAATCT) at approximately -80. This sequence is essential and is involved in binding transcription factors. Further upstream of the TATA box, eukaryotic promoters may also contain one or more GC-rich boxes (GGCG) or octamer boxes (ATTTGCAT). These elements bind cellular factors that increase the efficiency of transcription initiation and are often identified in more “active” genes that are constantly being expressed by the cell.
The complexity of eukaryotic transcription does not end with the polymerases and promoters. An army of basal transcription factors, enhancers, and silencers also help to regulate the frequency with which pre-mRNA is synthesized from a gene. Enhancers and silencers affect the efficiency of transcription but are not necessary for transcription to proceed. Basal transcription factors are crucial in the formation of a preinitiation complex on the DNA template that subsequently recruits RNA polymerase II for transcription initiation.
The names of the basal transcription factors begin with “TFII” (this is the transcription factor for RNA polymerase II) and are specified with the letters A–J. The transcription factors systematically fall into place on the DNA template, with each one further stabilizing the preinitiation complex and contributing to the recruitment of RNA polymerase II.
The processes of bringing RNA polymerases I and III to the DNA template involve slightly less complex collections of transcription factors, but the general theme is the same. Eukaryotic transcription is a tightly regulated process that requires a variety of proteins to interact with each other and with the DNA strand. Although the process of transcription in eukaryotes involves a greater metabolic investment than in prokaryotes, it ensures that the cell transcribes precisely the pre-mRNAs that it needs for protein synthesis.
The Evolution of PromotersThe evolution of genes may be a familiar concept. Mutations can occur in genes during DNA replication, and the result may or may not be beneficial to the cell. By altering an enzyme, structural protein, or some other factor, the process of mutation can transform functions or physical features. However, eukaryotic promoters and other gene regulatory sequences may evolve as well. For instance, consider a gene that, over many generations, becomes more valuable to the cell. Maybe the gene encodes a structural protein that the cell needs to synthesize in abundance for a certain function. If this is the case, it would be beneficial to the cell for that gene’s promoter to recruit transcription factors more efficiently and increase gene expression.
Scientists examining the evolution of promoter sequences have reported varying results. In part, this is because it is difficult to infer exactly where a eukaryotic promoter begins and ends. Some promoters occur within genes; others are located very far upstream, or even downstream, of the genes they are regulating. However, when researchers limited their examination to human core promoter sequences that were defined experimentally as sequences that bind the preinitiation complex, they found that promoters evolve even faster than protein-coding genes.
It is still unclear how promoter evolution might correspond to the evolution of humans or other higher organisms. However, the evolution of a promoter to effectively make more or less of a given gene product is an intriguing alternative to the evolution of the genes themselves.1
In eukaryotes, the conserved promoter elements differ for genes transcribed by RNA polymerases I, II, and III. RNA polymerase I transcribes genes that have two GC-rich promoter sequences in the -45 to +20 region. These sequences alone are sufficient for transcription initiation to occur, but promoters with additional sequences in the region from -180 to -105 upstream of the initiation site will further enhance initiation. Genes that are transcribed by RNA polymerase III have upstream promoters or promoters that occur within the genes themselves.
Following the formation of the preinitiation complex, the polymerase is released from the other transcription factors, and elongation is allowed to proceed as it does in prokaryotes with the polymerase synthesizing pre-mRNA in the 5' to 3' direction. As discussed previously, RNA polymerase II transcribes the major share of eukaryotic genes, so this section will focus on how this polymerase accomplishes elongation and termination.
Although the enzymatic process of elongation is essentially the same in eukaryotes and prokaryotes, the DNA template is more complex. When eukaryotic cells are not dividing, their genes exist as a diffuse mass of DNA and proteins called chromatin. The DNA is tightly packaged around charged histone proteins at repeated intervals. These DNA–histone complexes, collectively called nucleosomes, are regularly spaced and include 146 nucleotides of DNA wound around eight histones like thread around a spool.
For polynucleotide synthesis to occur, the transcription machinery needs to move histones out of the way every time it encounters a nucleosome. This is accomplished by a special protein complex called FACT, which stands for “facilitates chromatin transcription.” This complex pulls histones away from the DNA template as the polymerase moves along it. Once the pre-mRNA is synthesized, the FACT complex replaces the histones to recreate the nucleosomes.
The termination of transcription is different for the different polymerases. Unlike in prokaryotes, elongation by RNA polymerase II in eukaryotes takes place 1,000–2,000 nucleotides beyond the end of the gene being transcribed. This pre-mRNA tail is subsequently removed by cleavage during mRNA processing. On the other hand, RNA polymerases I and III require termination signals. Genes transcribed by RNA polymerase I contain a specific 18-nucleotide sequence that is recognized by a termination protein. The process of termination in RNA polymerase III involves an mRNA hairpin similar to rho-independent termination of transcription in prokaryotes.
Transcription in eukaryotes involves one of three types of polymerases, depending on the gene being transcribed. RNA polymerase II transcribes all of the protein-coding genes, whereas RNA polymerase I transcribes rRNA genes, and RNA polymerase III transcribes rRNA, tRNA, and small nuclear RNA genes. The initiation of transcription in eukaryotes involves the binding of several transcription factors to complex promoter sequences that are usually located upstream of the gene being copied. The mRNA is synthesized in the 5' to 3' direction, and the FACT complex moves and reassembles nucleosomes as the polymerase passes by. Whereas RNA polymerases I and III terminate transcription by protein- or RNA hairpin-dependent methods, RNA polymerase II transcribes for 1,000 or more nucleotides beyond the gene template and cleaves the excess during pre-mRNA processing.
[link] A scientist splices a eukaryotic promoter in front of a bacterial gene and inserts the gene in a bacterial chromosome. Would you expect the bacteria to transcribe the gene?
[link] No. Prokaryotes use different promoters than eukaryotes.
Which feature of promoters can be found in both prokaryotes and eukaryotes?
B
What transcripts will be most affected by low levels of α-amanitin?
B

You can also download for free at http://cnx.org/contents/185cbf87-c72e-48f5-b51e-f14f21b5eabd@11.2
Attribution: