By the end of this section, you will be able to:
The length of the cell cycle is highly variable, even within the cells of a single organism. In humans, the frequency of cell turnover ranges from a few hours in early embryonic development, to an average of two to five days for epithelial cells, and to an entire human lifetime spent in G0 by specialized cells, such as cortical neurons or cardiac muscle cells. There is also variation in the time that a cell spends in each phase of the cell cycle. When fast-dividing mammalian cells are grown in culture (outside the body under optimal growing conditions), the length of the cycle is about 24 hours. In rapidly dividing human cells with a 24-hour cell cycle, the G1 phase lasts approximately nine hours, the S phase lasts 10 hours, the G2 phase lasts about four and one-half hours, and the M phase lasts approximately one-half hour. In early embryos of fruit flies, the cell cycle is completed in about eight minutes. The timing of events in the cell cycle is controlled by mechanisms that are both internal and external to the cell.
Both the initiation and inhibition of cell division are triggered by events external to the cell when it is about to begin the replication process. An event may be as simple as the death of a nearby cell or as sweeping as the release of growth-promoting hormones, such as human growth hormone (HGH). A lack of HGH can inhibit cell division, resulting in dwarfism, whereas too much HGH can result in gigantism. Crowding of cells can also inhibit cell division. Another factor that can initiate cell division is the size of the cell; as a cell grows, it becomes inefficient due to its decreasing surface-to-volume ratio. The solution to this problem is to divide.
Whatever the source of the message, the cell receives the signal, and a series of events within the cell allows it to proceed into interphase. Moving forward from this initiation point, every parameter required during each cell cycle phase must be met or the cycle cannot progress.
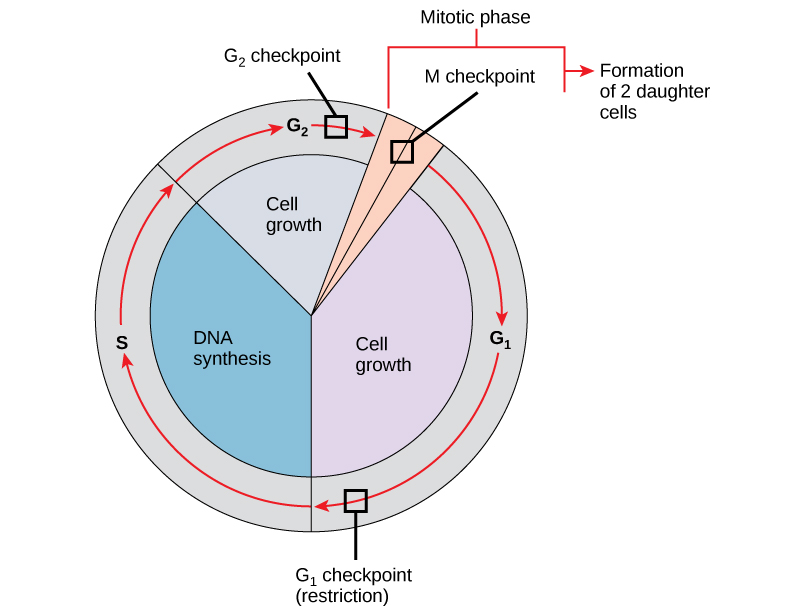
It is essential that the daughter cells produced be exact duplicates of the parent cell. Mistakes in the duplication or distribution of the chromosomes lead to mutations that may be passed forward to every new cell produced from an abnormal cell. To prevent a compromised cell from continuing to divide, there are internal control mechanisms that operate at three main cell cycle checkpoints. A checkpoint is one of several points in the eukaryotic cell cycle at which the progression of a cell to the next stage in the cycle can be halted until conditions are favorable. These checkpoints occur near the end of G1, at the G2/M transition, and during metaphase ([link]).

The G1 checkpoint determines whether all conditions are favorable for cell division to proceed. The G1 checkpoint, also called the restriction point (in yeast), is a point at which the cell irreversibly commits to the cell division process. External influences, such as growth factors, play a large role in carrying the cell past the G1 checkpoint. In addition to adequate reserves and cell size, there is a check for genomic DNA damage at the G1 checkpoint. A cell that does not meet all the requirements will not be allowed to progress into the S phase. The cell can halt the cycle and attempt to remedy the problematic condition, or the cell can advance into G0 and await further signals when conditions improve.
The G2 checkpoint bars entry into the mitotic phase if certain conditions are not met. As at the G1 checkpoint, cell size and protein reserves are assessed. However, the most important role of the G2 checkpoint is to ensure that all of the chromosomes have been replicated and that the replicated DNA is not damaged. If the checkpoint mechanisms detect problems with the DNA, the cell cycle is halted, and the cell attempts to either complete DNA replication or repair the damaged DNA.
The M checkpoint occurs near the end of the metaphase stage of karyokinesis. The M checkpoint is also known as the spindle checkpoint, because it determines whether all the sister chromatids are correctly attached to the spindle microtubules. Because the separation of the sister chromatids during anaphase is an irreversible step, the cycle will not proceed until the kinetochores of each pair of sister chromatids are firmly anchored to at least two spindle fibers arising from opposite poles of the cell.
 Watch what occurs at the G1, G2, and M checkpoints by visiting this website to see an animation of the cell cycle.
Watch what occurs at the G1, G2, and M checkpoints by visiting this website to see an animation of the cell cycle.
In addition to the internally controlled checkpoints, there are two groups of intracellular molecules that regulate the cell cycle. These regulatory molecules either promote progress of the cell to the next phase (positive regulation) or halt the cycle (negative regulation). Regulator molecules may act individually, or they can influence the activity or production of other regulatory proteins. Therefore, the failure of a single regulator may have almost no effect on the cell cycle, especially if more than one mechanism controls the same event. Conversely, the effect of a deficient or non-functioning regulator can be wide-ranging and possibly fatal to the cell if multiple processes are affected.
Two groups of proteins, called cyclins and cyclin-dependent kinases (Cdks), are responsible for the progress of the cell through the various checkpoints. The levels of the four cyclin proteins fluctuate throughout the cell cycle in a predictable pattern ([link]). Increases in the concentration of cyclin proteins are triggered by both external and internal signals. After the cell moves to the next stage of the cell cycle, the cyclins that were active in the previous stage are degraded.
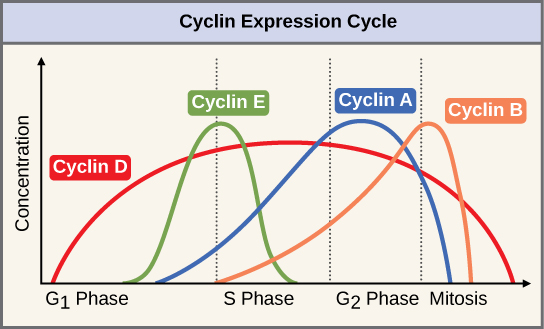
Cyclins regulate the cell cycle only when they are tightly bound to Cdks. To be fully active, the Cdk/cyclin complex must also be phosphorylated in specific locations. Like all kinases, Cdks are enzymes (kinases) that phosphorylate other proteins. Phosphorylation activates the protein by changing its shape. The proteins phosphorylated by Cdks are involved in advancing the cell to the next phase. ([link]). The levels of Cdk proteins are relatively stable throughout the cell cycle; however, the concentrations of cyclin fluctuate and determine when Cdk/cyclin complexes form. The different cyclins and Cdks bind at specific points in the cell cycle and thus regulate different checkpoints.
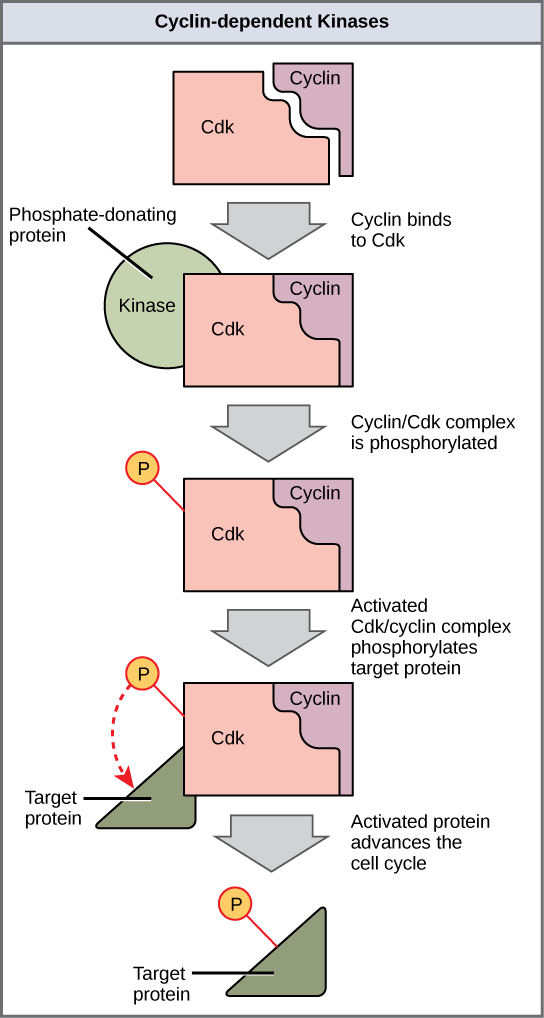
Since the cyclic fluctuations of cyclin levels are based on the timing of the cell cycle and not on specific events, regulation of the cell cycle usually occurs by either the Cdk molecules alone or the Cdk/cyclin complexes. Without a specific concentration of fully activated cyclin/Cdk complexes, the cell cycle cannot proceed through the checkpoints.
Although the cyclins are the main regulatory molecules that determine the forward momentum of the cell cycle, there are several other mechanisms that fine-tune the progress of the cycle with negative, rather than positive, effects. These mechanisms essentially block the progression of the cell cycle until problematic conditions are resolved. Molecules that prevent the full activation of Cdks are called Cdk inhibitors. Many of these inhibitor molecules directly or indirectly monitor a particular cell cycle event. The block placed on Cdks by inhibitor molecules will not be removed until the specific event that the inhibitor monitors is completed.
The second group of cell cycle regulatory molecules are negative regulators. Negative regulators halt the cell cycle. Remember that in positive regulation, active molecules cause the cycle to progress.
The best understood negative regulatory molecules are retinoblastoma protein (Rb), p53, and p21. Retinoblastoma proteins are a group of tumor-suppressor proteins common in many cells. The 53 and 21 designations refer to the functional molecular masses of the proteins (p) in kilodaltons. Much of what is known about cell cycle regulation comes from research conducted with cells that have lost regulatory control. All three of these regulatory proteins were discovered to be damaged or non-functional in cells that had begun to replicate uncontrollably (became cancerous). In each case, the main cause of the unchecked progress through the cell cycle was a faulty copy of the regulatory protein.
Rb, p53, and p21 act primarily at the G1 checkpoint. p53 is a multi-functional protein that has a major impact on the commitment of a cell to division because it acts when there is damaged DNA in cells that are undergoing the preparatory processes during G1. If damaged DNA is detected, p53 halts the cell cycle and recruits enzymes to repair the DNA. If the DNA cannot be repaired, p53 can trigger apoptosis, or cell suicide, to prevent the duplication of damaged chromosomes. As p53 levels rise, the production of p21 is triggered. p21 enforces the halt in the cycle dictated by p53 by binding to and inhibiting the activity of the Cdk/cyclin complexes. As a cell is exposed to more stress, higher levels of p53 and p21 accumulate, making it less likely that the cell will move into the S phase.
Rb exerts its regulatory influence on other positive regulator proteins. Chiefly, Rb monitors cell size. In the active, dephosphorylated state, Rb binds to proteins called transcription factors, most commonly, E2F ([link]). Transcription factors “turn on” specific genes, allowing the production of proteins encoded by that gene. When Rb is bound to E2F, production of proteins necessary for the G1/S transition is blocked. As the cell increases in size, Rb is slowly phosphorylated until it becomes inactivated. Rb releases E2F, which can now turn on the gene that produces the transition protein, and this particular block is removed. For the cell to move past each of the checkpoints, all positive regulators must be “turned on,” and all negative regulators must be “turned off.”
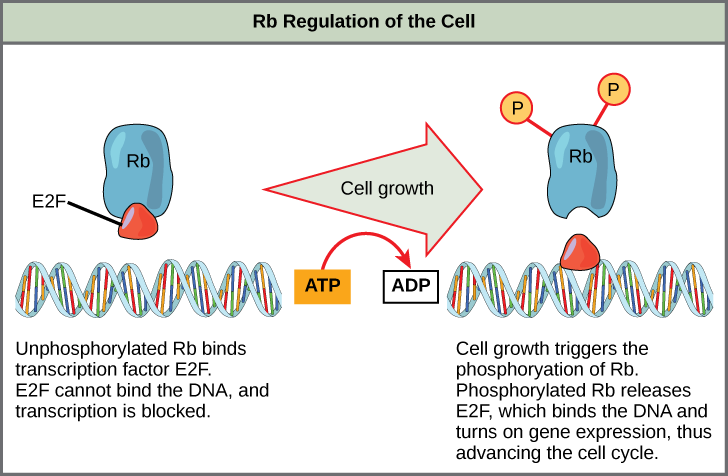
Rb and other proteins that negatively regulate the cell cycle are sometimes called tumor suppressors. Why do you think the name tumor suppressor might be appropriate for these proteins?
Each step of the cell cycle is monitored by internal controls called checkpoints. There are three major checkpoints in the cell cycle: one near the end of G1, a second at the G2/M transition, and the third during metaphase. Positive regulator molecules allow the cell cycle to advance to the next stage. Negative regulator molecules monitor cellular conditions and can halt the cycle until specific requirements are met.
[link] Rb and other proteins that negatively regulate the cell cycle are sometimes called tumor suppressors. Why do you think the name tumor suppressor might be an appropriate for these proteins?
[link] Rb and other negative regulatory proteins control cell division and therefore prevent the formation of tumors. Mutations that prevent these proteins from carrying out their function can result in cancer.
At which of the cell cycle checkpoints do external forces have the greatest influence?
A
What is the main prerequisite for clearance at the G2 checkpoint?
C
If the M checkpoint is not cleared, what stage of mitosis will be blocked?
D
Which protein is a positive regulator that phosphorylates other proteins when activated?
D
Many of the negative regulator proteins of the cell cycle were discovered in what type of cells?
C
Which negative regulatory molecule can trigger cell suicide (apoptosis) if vital cell cycle events do not occur?
A
Describe the general conditions that must be met at each of the three main cell cycle checkpoints.
The G1 checkpoint monitors adequate cell growth, the state of the genomic DNA, adequate stores of energy, and materials for S phase. At the G2 checkpoint, DNA is checked to ensure that all chromosomes were duplicated and that there are no mistakes in newly synthesized DNA. Additionally, cell size and energy reserves are evaluated. The M checkpoint confirms the correct attachment of the mitotic spindle fibers to the kinetochores.
Explain the roles of the positive cell cycle regulators compared to the negative regulators.
Positive cell regulators such as cyclin and Cdk perform tasks that advance the cell cycle to the next stage. Negative regulators such as Rb, p53, and p21 block the progression of the cell cycle until certain events have occurred.
What steps are necessary for Cdk to become fully active?
Cdk must bind to a cyclin, and it must be phosphorylated in the correct position to become fully active.
Rb is a negative regulator that blocks the cell cycle at the G1 checkpoint until the cell achieves a requisite size. What molecular mechanism does Rb employ to halt the cell cycle?
Rb is active when it is dephosphorylated. In this state, Rb binds to E2F, which is a transcription factor required for the transcription and eventual translation of molecules required for the G1/S transition. E2F cannot transcribe certain genes when it is bound to Rb. As the cell increases in size, Rb becomes phosphorylated, inactivated, and releases E2F. E2F can then promote the transcription of the genes it controls, and the transition proteins will be produced.

You can also download for free at http://cnx.org/contents/185cbf87-c72e-48f5-b51e-f14f21b5eabd@11.2
Attribution: