You isolate a cell strain in which the joining together of Okazaki fragments is impaired and suspect that a mutation has occurred in an enzyme found at the replication fork. Which enzyme is most likely to be mutated?
By the end of this section, you will be able to:
DNA replication has been extremely well studied in prokaryotes primarily because of the small size of the genome and the mutants that are available. E. coli has 4.6 million base pairs in a single circular chromosome and all of it gets replicated in approximately 42 minutes, starting from a single origin of replication and proceeding around the circle in both directions. This means that approximately 1000 nucleotides are added per second. The process is quite rapid and occurs without many mistakes.
DNA replication employs a large number of proteins and enzymes, each of which plays a critical role during the process. One of the key players is the enzyme DNA polymerase, also known as DNA pol, which adds nucleotides one by one to the growing DNA chain that are complementary to the template strand. The addition of nucleotides requires energy; this energy is obtained from the nucleotides that have three phosphates attached to them, similar to ATP which has three phosphate groups attached. When the bond between the phosphates is broken, the energy released is used to form the phosphodiester bond between the incoming nucleotide and the growing chain. In prokaryotes, three main types of polymerases are known: DNA pol I, DNA pol II, and DNA pol III. It is now known that DNA pol III is the enzyme required for DNA synthesis; DNA pol I and DNA pol II are primarily required for repair.
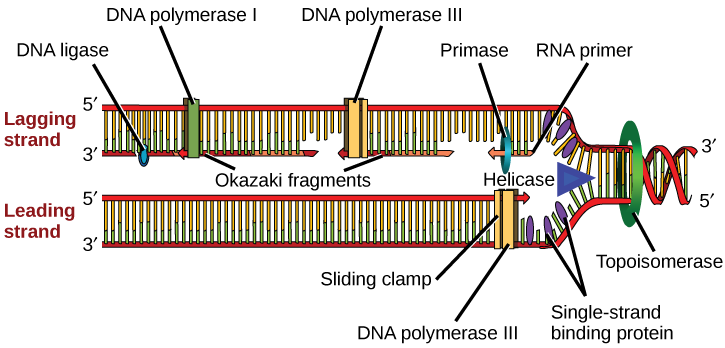
How does the replication machinery know where to begin? It turns out that there are specific nucleotide sequences called origins of replication where replication begins. In E. coli, which has a single origin of replication on its one chromosome (as do most prokaryotes), it is approximately 245 base pairs long and is rich in AT sequences. The origin of replication is recognized by certain proteins that bind to this site. An enzyme called helicase unwinds the DNA by breaking the hydrogen bonds between the nitrogenous base pairs. ATP hydrolysis is required for this process. As the DNA opens up, Y-shaped structures called replication forks are formed. Two replication forks are formed at the origin of replication and these get extended bi- directionally as replication proceeds. Single-strand binding proteins coat the single strands of DNA near the replication fork to prevent the single-stranded DNA from winding back into a double helix. DNA polymerase is able to add nucleotides only in the 5' to 3' direction (a new DNA strand can be only extended in this direction). It also requires a free 3'-OH group to which it can add nucleotides by forming a phosphodiester bond between the 3'-OH end and the 5' phosphate of the next nucleotide. This essentially means that it cannot add nucleotides if a free 3'-OH group is not available. Then how does it add the first nucleotide? The problem is solved with the help of a primer that provides the free 3'-OH end. Another enzyme, RNA primase, synthesizes an RNA primer that is about five to ten nucleotides long and complementary to the DNA. Because this sequence primes the DNA synthesis, it is appropriately called the primer. DNA polymerase can now extend this RNA primer, adding nucleotides one by one that are complementary to the template strand ([link]).

You isolate a cell strain in which the joining together of Okazaki fragments is impaired and suspect that a mutation has occurred in an enzyme found at the replication fork. Which enzyme is most likely to be mutated?
The replication fork moves at the rate of 1000 nucleotides per second. DNA polymerase can only extend in the 5' to 3' direction, which poses a slight problem at the replication fork. As we know, the DNA double helix is anti-parallel; that is, one strand is in the 5' to 3' direction and the other is oriented in the 3' to 5' direction. One strand, which is complementary to the 3' to 5' parental DNA strand, is synthesized continuously towards the replication fork because the polymerase can add nucleotides in this direction. This continuously synthesized strand is known as the leading strand. The other strand, complementary to the 5' to 3' parental DNA, is extended away from the replication fork, in small fragments known as Okazaki fragments, each requiring a primer to start the synthesis. Okazaki fragments are named after the Japanese scientist who first discovered them. The strand with the Okazaki fragments is known as the lagging strand.
The leading strand can be extended by one primer alone, whereas the lagging strand needs a new primer for each of the short Okazaki fragments. The overall direction of the lagging strand will be 3' to 5', and that of the leading strand 5' to 3'. A protein called the sliding clamp holds the DNA polymerase in place as it continues to add nucleotides. The sliding clamp is a ring-shaped protein that binds to the DNA and holds the polymerase in place. Topoisomerase prevents the over-winding of the DNA double helix ahead of the replication fork as the DNA is opening up; it does so by causing temporary nicks in the DNA helix and then resealing it. As synthesis proceeds, the RNA primers are replaced by DNA. The primers are removed by the exonuclease activity of DNA pol I, and the gaps are filled in by deoxyribonucleotides. The nicks that remain between the newly synthesized DNA (that replaced the RNA primer) and the previously synthesized DNA are sealed by the enzyme DNA ligase that catalyzes the formation of phosphodiester linkage between the 3'-OH end of one nucleotide and the 5' phosphate end of the other fragment.
Once the chromosome has been completely replicated, the two DNA copies move into two different cells during cell division. The process of DNA replication can be summarized as follows:
[link] summarizes the enzymes involved in prokaryotic DNA replication and the functions of each.
| Prokaryotic DNA Replication: Enzymes and Their Function | |
|---|---|
| Enzyme/protein | Specific Function |
| DNA pol I | Exonuclease activity removes RNA primer and replaces with newly synthesized DNA |
| DNA pol II | Repair function |
| DNA pol III | Main enzyme that adds nucleotides in the 5'-3' direction |
| Helicase | Opens the DNA helix by breaking hydrogen bonds between the nitrogenous bases |
| Ligase | Seals the gaps between the Okazaki fragments to create one continuous DNA strand |
| Primase | Synthesizes RNA primers needed to start replication |
| Sliding Clamp | Helps to hold the DNA polymerase in place when nucleotides are being added |
| Topoisomerase | Helps relieve the stress on DNA when unwinding by causing breaks and then resealing the DNA |
| Single-strand binding proteins (SSB) | Binds to single-stranded DNA to avoid DNA rewinding back. |
 Review the full process of DNA replication here.
Review the full process of DNA replication here.
Replication in prokaryotes starts from a sequence found on the chromosome called the origin of replication—the point at which the DNA opens up. Helicase opens up the DNA double helix, resulting in the formation of the replication fork. Single-strand binding proteins bind to the single-stranded DNA near the replication fork to keep the fork open. Primase synthesizes an RNA primer to initiate synthesis by DNA polymerase, which can add nucleotides only in the 5' to 3' direction. One strand is synthesized continuously in the direction of the replication fork; this is called the leading strand. The other strand is synthesized in a direction away from the replication fork, in short stretches of DNA known as Okazaki fragments. This strand is known as the lagging strand. Once replication is completed, the RNA primers are replaced by DNA nucleotides and the DNA is sealed with DNA ligase, which creates phosphodiester bonds between the 3'-OH of one end and the 5' phosphate of the other strand.
[link] You isolate a cell strain in which the joining together of Okazaki fragments is impaired and suspect that a mutation has occurred in an enzyme found at the replication fork. Which enzyme is most likely to be mutated?
[link] DNA ligase, as this enzyme joins together Okazaki fragments.
Which of the following components is not involved during the formation of the replication fork?
D
Which of the following does the enzyme primase synthesize?
B
In which direction does DNA replication take place?
A
DNA replication is bidirectional and discontinuous; explain your understanding of those concepts.
At an origin of replication, two replication forks are formed that are extended in two directions. On the lagging strand, Okazaki fragments are formed in a discontinuous manner.
What are Okazaki fragments and how they are formed?
Short DNA fragments are formed on the lagging strand synthesized in a direction away from the replication fork. These are synthesized by DNA pol.
If the rate of replication in a particular prokaryote is 900 nucleotides per second, how long would it take 1.2 million base pair genomes to make two copies?
1333 seconds or 22.2 minutes.
Explain the events taking place at the replication fork. If the gene for helicase is mutated, what part of replication will be affected?
At the replication fork, the events taking place are helicase action, binding of single-strand binding proteins, primer synthesis, and synthesis of new strands. If there is a mutated helicase gene, the replication fork will not be extended.
What is the role of a primer in DNA replication? What would happen if you forgot to add a primer in a tube containing the reaction mix for a DNA sequencing reaction?
Primer provides a 3’-OH group for DNA pol to start adding nucleotides. There would be no reaction in the tube without a primer, and no bands would be visible on the electrophoresis.

You can also download for free at http://cnx.org/contents/185cbf87-c72e-48f5-b51e-f14f21b5eabd@11.2
Attribution: