
By the end of this section, you will be able to:
Mendel generalized the results of his pea-plant experiments into four postulates, some of which are sometimes called “laws,” that describe the basis of dominant and recessive inheritance in diploid organisms. As you have learned, more complex extensions of Mendelism exist that do not exhibit the same F2 phenotypic ratios (3:1). Nevertheless, these laws summarize the basics of classical genetics.
Mendel proposed first that paired unit factors of heredity were transmitted faithfully from generation to generation by the dissociation and reassociation of paired factors during gametogenesis and fertilization, respectively. After he crossed peas with contrasting traits and found that the recessive trait resurfaced in the F2 generation, Mendel deduced that hereditary factors must be inherited as discrete units. This finding contradicted the belief at that time that parental traits were blended in the offspring.
Mendel’s law of dominance states that in a heterozygote, one trait will conceal the presence of another trait for the same characteristic. Rather than both alleles contributing to a phenotype, the dominant allele will be expressed exclusively. The recessive allele will remain “latent” but will be transmitted to offspring by the same manner in which the dominant allele is transmitted. The recessive trait will only be expressed by offspring that have two copies of this allele ([link]), and these offspring will breed true when self-crossed.
Since Mendel’s experiments with pea plants, other researchers have found that the law of dominance does not always hold true. Instead, several different patterns of inheritance have been found to exist.

Observing that true-breeding pea plants with contrasting traits gave rise to F1 generations that all expressed the dominant trait and F2 generations that expressed the dominant and recessive traits in a 3:1 ratio, Mendel proposed the law of segregation. This law states that paired unit factors (genes) must segregate equally into gametes such that offspring have an equal likelihood of inheriting either factor. For the F2 generation of a monohybrid cross, the following three possible combinations of genotypes could result: homozygous dominant, heterozygous, or homozygous recessive. Because heterozygotes could arise from two different pathways (receiving one dominant and one recessive allele from either parent), and because heterozygotes and homozygous dominant individuals are phenotypically identical, the law supports Mendel’s observed 3:1 phenotypic ratio. The equal segregation of alleles is the reason we can apply the Punnett square to accurately predict the offspring of parents with known genotypes. The physical basis of Mendel’s law of segregation is the first division of meiosis, in which the homologous chromosomes with their different versions of each gene are segregated into daughter nuclei. The role of the meiotic segregation of chromosomes in sexual reproduction was not understood by the scientific community during Mendel’s lifetime.
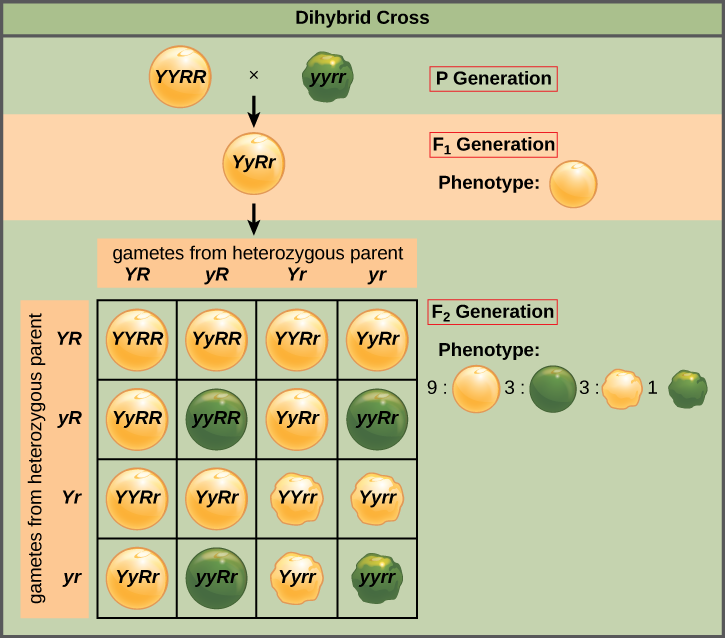
Mendel’s law of independent assortment states that genes do not influence each other with regard to the sorting of alleles into gametes, and every possible combination of alleles for every gene is equally likely to occur. The independent assortment of genes can be illustrated by the dihybrid cross, a cross between two true-breeding parents that express different traits for two characteristics. Consider the characteristics of seed color and seed texture for two pea plants, one that has green, wrinkled seeds (yyrr) and another that has yellow, round seeds (YYRR). Because each parent is homozygous, the law of segregation indicates that the gametes for the green/wrinkled plant all are yr, and the gametes for the yellow/round plant are all YR. Therefore, the F1 generation of offspring all are YyRr ([link]).
In pea plants, purple flowers (P) are dominant to white flowers (p) and yellow peas (Y) are dominant to green peas (y). What are the possible genotypes and phenotypes for a cross between PpYY and ppYy pea plants? How many squares do you need to do a Punnett square analysis of this cross?
For the F2 generation, the law of segregation requires that each gamete receive either an R allele or an r allele along with either a Y allele or a y allele. The law of independent assortment states that a gamete into which an r allele sorted would be equally likely to contain either a Y allele or a y allele. Thus, there are four equally likely gametes that can be formed when the YyRr heterozygote is self-crossed, as follows: YR, Yr, yR, and yr. Arranging these gametes along the top and left of a 4 × 4 Punnett square ([link]) gives us 16 equally likely genotypic combinations. From these genotypes, we infer a phenotypic ratio of 9 round/yellow:3 round/green:3 wrinkled/yellow:1 wrinkled/green ([link]). These are the offspring ratios we would expect, assuming we performed the crosses with a large enough sample size.
Because of independent assortment and dominance, the 9:3:3:1 dihybrid phenotypic ratio can be collapsed into two 3:1 ratios, characteristic of any monohybrid cross that follows a dominant and recessive pattern. Ignoring seed color and considering only seed texture in the above dihybrid cross, we would expect that three quarters of the F2 generation offspring would be round, and one quarter would be wrinkled. Similarly, isolating only seed color, we would assume that three quarters of the F2 offspring would be yellow and one quarter would be green. The sorting of alleles for texture and color are independent events, so we can apply the product rule. Therefore, the proportion of round and yellow F2 offspring is expected to be (3/4) × (3/4) = 9/16, and the proportion of wrinkled and green offspring is expected to be (1/4) × (1/4) = 1/16. These proportions are identical to those obtained using a Punnett square. Round, green and wrinkled, yellow offspring can also be calculated using the product rule, as each of these genotypes includes one dominant and one recessive phenotype. Therefore, the proportion of each is calculated as (3/4) × (1/4) = 3/16.
The law of independent assortment also indicates that a cross between yellow, wrinkled (YYrr) and green, round (yyRR) parents would yield the same F1 and F2 offspring as in the YYRR x yyrr cross.
The physical basis for the law of independent assortment also lies in meiosis I, in which the different homologous pairs line up in random orientations. Each gamete can contain any combination of paternal and maternal chromosomes (and therefore the genes on them) because the orientation of tetrads on the metaphase plane is random.
When more than two genes are being considered, the Punnett-square method becomes unwieldy. For instance, examining a cross involving four genes would require a 16 × 16 grid containing 256 boxes. It would be extremely cumbersome to manually enter each genotype. For more complex crosses, the forked-line and probability methods are preferred.
To prepare a forked-line diagram for a cross between F1 heterozygotes resulting from a cross between AABBCC and aabbcc parents, we first create rows equal to the number of genes being considered, and then segregate the alleles in each row on forked lines according to the probabilities for individual monohybrid crosses ([link]). We then multiply the values along each forked path to obtain the F2 offspring probabilities. Note that this process is a diagrammatic version of the product rule. The values along each forked pathway can be multiplied because each gene assorts independently. For a trihybrid cross, the F2 phenotypic ratio is 27:9:9:9:3:3:3:1.
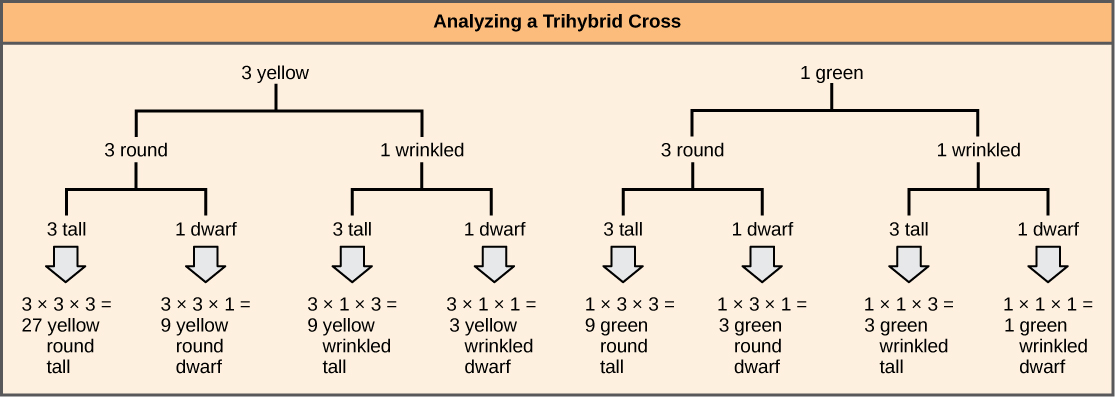
While the forked-line method is a diagrammatic approach to keeping track of probabilities in a cross, the probability method gives the proportions of offspring expected to exhibit each phenotype (or genotype) without the added visual assistance. Both methods make use of the product rule and consider the alleles for each gene separately. Earlier, we examined the phenotypic proportions for a trihybrid cross using the forked-line method; now we will use the probability method to examine the genotypic proportions for a cross with even more genes.
For a trihybrid cross, writing out the forked-line method is tedious, albeit not as tedious as using the Punnett-square method. To fully demonstrate the power of the probability method, however, we can consider specific genetic calculations. For instance, for a tetrahybrid cross between individuals that are heterozygotes for all four genes, and in which all four genes are sorting independently and in a dominant and recessive pattern, what proportion of the offspring will be expected to be homozygous recessive for all four alleles? Rather than writing out every possible genotype, we can use the probability method. We know that for each gene, the fraction of homozygous recessive offspring will be 1/4. Therefore, multiplying this fraction for each of the four genes, (1/4) × (1/4) × (1/4) × (1/4), we determine that 1/256 of the offspring will be quadruply homozygous recessive.
For the same tetrahybrid cross, what is the expected proportion of offspring that have the dominant phenotype at all four loci? We can answer this question using phenotypic proportions, but let’s do it the hard way—using genotypic proportions. The question asks for the proportion of offspring that are 1) homozygous dominant at A or heterozygous at A, and 2) homozygous at B or heterozygous at B, and so on. Noting the “or” and “and” in each circumstance makes clear where to apply the sum and product rules. The probability of a homozygous dominant at A is 1/4 and the probability of a heterozygote at A is 1/2. The probability of the homozygote or the heterozygote is 1/4 + 1/2 = 3/4 using the sum rule. The same probability can be obtained in the same way for each of the other genes, so that the probability of a dominant phenotype at A and B and C and D is, using the product rule, equal to 3/4 × 3/4 × 3/4 × 3/4, or 27/64. If you are ever unsure about how to combine probabilities, returning to the forked-line method should make it clear.
Predicting the genotypes and phenotypes of offspring from given crosses is the best way to test your knowledge of Mendelian genetics. Given a multihybrid cross that obeys independent assortment and follows a dominant and recessive pattern, several generalized rules exist; you can use these rules to check your results as you work through genetics calculations ([link]). To apply these rules, first you must determine n, the number of heterozygous gene pairs (the number of genes segregating two alleles each). For example, a cross between AaBb and AaBb heterozygotes has an n of 2. In contrast, a cross between AABb and AABb has an n of 1 because A is not heterozygous.
| General Rules for Multihybrid Crosses | |
|---|---|
| General Rule | Number of Heterozygous Gene Pairs |
| Number of different F1 gametes | 2n |
| Number of different F2 genotypes | 3n |
| Given dominant and recessive inheritance, the number of different F2 phenotypes | 2n |
Although all of Mendel’s pea characteristics behaved according to the law of independent assortment, we now know that some allele combinations are not inherited independently of each other. Genes that are located on separate non-homologous chromosomes will always sort independently. However, each chromosome contains hundreds or thousands of genes, organized linearly on chromosomes like beads on a string. The segregation of alleles into gametes can be influenced by linkage, in which genes that are located physically close to each other on the same chromosome are more likely to be inherited as a pair. However, because of the process of recombination, or “crossover,” it is possible for two genes on the same chromosome to behave independently, or as if they are not linked. To understand this, let’s consider the biological basis of gene linkage and recombination.
Homologous chromosomes possess the same genes in the same linear order. The alleles may differ on homologous chromosome pairs, but the genes to which they correspond do not. In preparation for the first division of meiosis, homologous chromosomes replicate and synapse. Like genes on the homologs align with each other. At this stage, segments of homologous chromosomes exchange linear segments of genetic material ([link]). This process is called recombination, or crossover, and it is a common genetic process. Because the genes are aligned during recombination, the gene order is not altered. Instead, the result of recombination is that maternal and paternal alleles are combined onto the same chromosome. Across a given chromosome, several recombination events may occur, causing extensive shuffling of alleles.
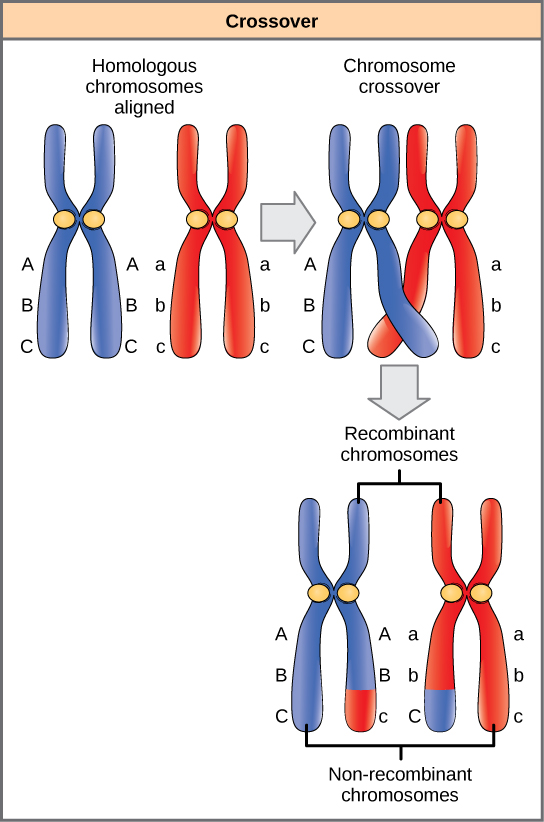
When two genes are located in close proximity on the same chromosome, they are considered linked, and their alleles tend to be transmitted through meiosis together. To exemplify this, imagine a dihybrid cross involving flower color and plant height in which the genes are next to each other on the chromosome. If one homologous chromosome has alleles for tall plants and red flowers, and the other chromosome has genes for short plants and yellow flowers, then when the gametes are formed, the tall and red alleles will go together into a gamete and the short and yellow alleles will go into other gametes. These are called the parental genotypes because they have been inherited intact from the parents of the individual producing gametes. But unlike if the genes were on different chromosomes, there will be no gametes with tall and yellow alleles and no gametes with short and red alleles. If you create the Punnett square with these gametes, you will see that the classical Mendelian prediction of a 9:3:3:1 outcome of a dihybrid cross would not apply. As the distance between two genes increases, the probability of one or more crossovers between them increases, and the genes behave more like they are on separate chromosomes. Geneticists have used the proportion of recombinant gametes (the ones not like the parents) as a measure of how far apart genes are on a chromosome. Using this information, they have constructed elaborate maps of genes on chromosomes for well-studied organisms, including humans.
Mendel’s seminal publication makes no mention of linkage, and many researchers have questioned whether he encountered linkage but chose not to publish those crosses out of concern that they would invalidate his independent assortment postulate. The garden pea has seven chromosomes, and some have suggested that his choice of seven characteristics was not a coincidence. However, even if the genes he examined were not located on separate chromosomes, it is possible that he simply did not observe linkage because of the extensive shuffling effects of recombination.
Testing the Hypothesis of Independent AssortmentTo better appreciate the amount of labor and ingenuity that went into Mendel’s experiments, proceed through one of Mendel’s dihybrid crosses.
Question: What will be the offspring of a dihybrid cross?
Background: Consider that pea plants mature in one growing season, and you have access to a large garden in which you can cultivate thousands of pea plants. There are several true-breeding plants with the following pairs of traits: tall plants with inflated pods, and dwarf plants with constricted pods. Before the plants have matured, you remove the pollen-producing organs from the tall/inflated plants in your crosses to prevent self-fertilization. Upon plant maturation, the plants are manually crossed by transferring pollen from the dwarf/constricted plants to the stigmata of the tall/inflated plants.
Hypothesis: Both trait pairs will sort independently according to Mendelian laws. When the true-breeding parents are crossed, all of the F1 offspring are tall and have inflated pods, which indicates that the tall and inflated traits are dominant over the dwarf and constricted traits, respectively. A self-cross of the F1 heterozygotes results in 2,000 F2 progeny.
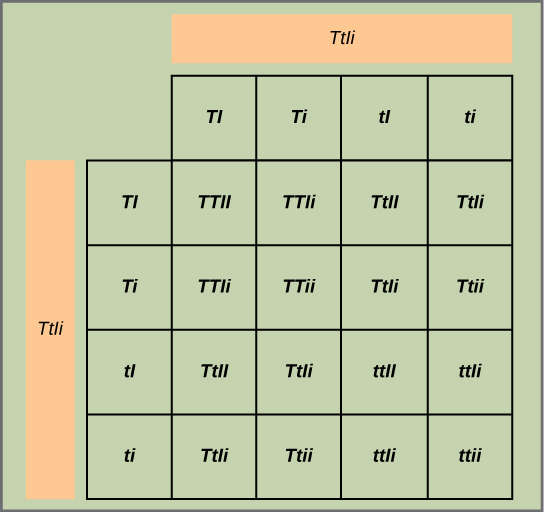
Test the hypothesis: Because each trait pair sorts independently, the ratios of tall:dwarf and inflated:constricted are each expected to be 3:1. The tall/dwarf trait pair is called T/t, and the inflated/constricted trait pair is designated I/i. Each member of the F1 generation therefore has a genotype of TtIi. Construct a grid analogous to [link], in which you cross two TtIi individuals. Each individual can donate four combinations of two traits: TI, Ti, tI, or ti, meaning that there are 16 possibilities of offspring genotypes. Because the T and I alleles are dominant, any individual having one or two of those alleles will express the tall or inflated phenotypes, respectively, regardless if they also have a t or i allele. Only individuals that are tt or ii will express the dwarf and constricted alleles, respectively. As shown in [link], you predict that you will observe the following offspring proportions: tall/inflated:tall/constricted:dwarf/inflated:dwarf/constricted in a 9:3:3:1 ratio. Notice from the grid that when considering the tall/dwarf and inflated/constricted trait pairs in isolation, they are each inherited in 3:1 ratios.
Test the hypothesis: You cross the dwarf and tall plants and then self-cross the offspring. For best results, this is repeated with hundreds or even thousands of pea plants. What special precautions should be taken in the crosses and in growing the plants?
Analyze your data: You observe the following plant phenotypes in the F2 generation: 2706 tall/inflated, 930 tall/constricted, 888 dwarf/inflated, and 300 dwarf/constricted. Reduce these findings to a ratio and determine if they are consistent with Mendelian laws.
Form a conclusion: Were the results close to the expected 9:3:3:1 phenotypic ratio? Do the results support the prediction? What might be observed if far fewer plants were used, given that alleles segregate randomly into gametes? Try to imagine growing that many pea plants, and consider the potential for experimental error. For instance, what would happen if it was extremely windy one day?
Mendel’s studies in pea plants implied that the sum of an individual’s phenotype was controlled by genes (or as he called them, unit factors), such that every characteristic was distinctly and completely controlled by a single gene. In fact, single observable characteristics are almost always under the influence of multiple genes (each with two or more alleles) acting in unison. For example, at least eight genes contribute to eye color in humans.
 Eye color in humans is determined by multiple genes. Use the Eye Color Calculator to predict the eye color of children from parental eye color.
Eye color in humans is determined by multiple genes. Use the Eye Color Calculator to predict the eye color of children from parental eye color.
In some cases, several genes can contribute to aspects of a common phenotype without their gene products ever directly interacting. In the case of organ development, for instance, genes may be expressed sequentially, with each gene adding to the complexity and specificity of the organ. Genes may function in complementary or synergistic fashions, such that two or more genes need to be expressed simultaneously to affect a phenotype. Genes may also oppose each other, with one gene modifying the expression of another.
In epistasis, the interaction between genes is antagonistic, such that one gene masks or interferes with the expression of another. “Epistasis” is a word composed of Greek roots that mean “standing upon.” The alleles that are being masked or silenced are said to be hypostatic to the epistatic alleles that are doing the masking. Often the biochemical basis of epistasis is a gene pathway in which the expression of one gene is dependent on the function of a gene that precedes or follows it in the pathway.
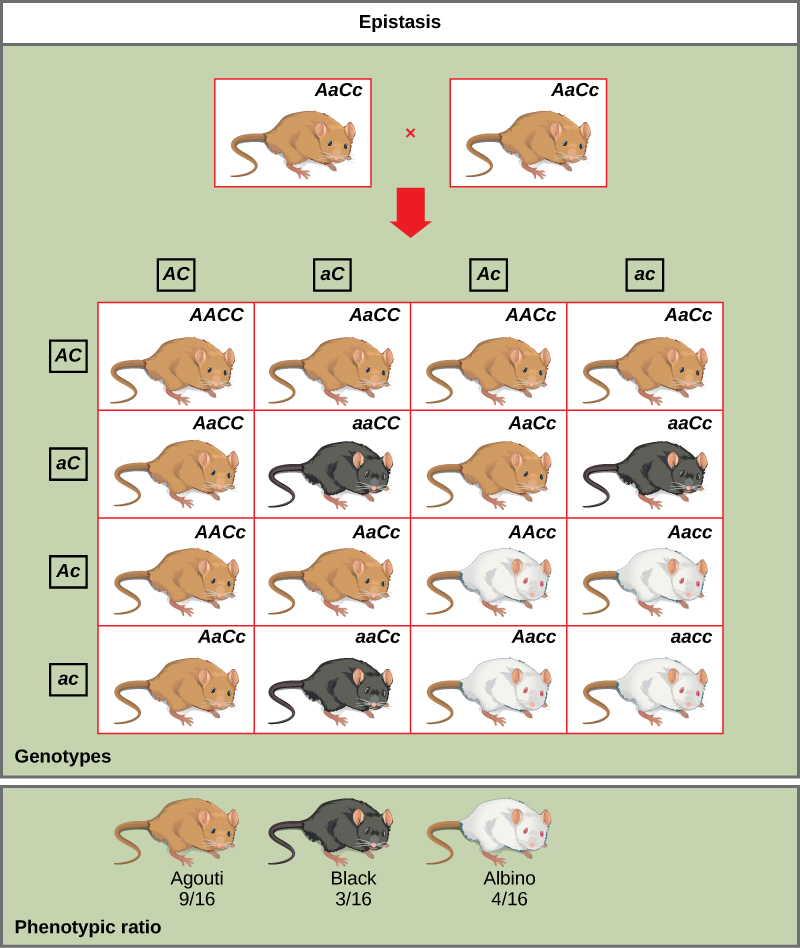
An example of epistasis is pigmentation in mice. The wild-type coat color, agouti (AA), is dominant to solid-colored fur (aa). However, a separate gene (C) is necessary for pigment production. A mouse with a recessive c allele at this locus is unable to produce pigment and is albino regardless of the allele present at locus A ([link]). Therefore, the genotypes AAcc, Aacc, and aacc all produce the same albino phenotype. A cross between heterozygotes for both genes (AaCc x AaCc) would generate offspring with a phenotypic ratio of 9 agouti:3 solid color:4 albino ([link]). In this case, the C gene is epistatic to the A gene.
Epistasis can also occur when a dominant allele masks expression at a separate gene. Fruit color in summer squash is expressed in this way. Homozygous recessive expression of the W gene (ww) coupled with homozygous dominant or heterozygous expression of the Y gene (YY or Yy) generates yellow fruit, and the wwyy genotype produces green fruit. However, if a dominant copy of the W gene is present in the homozygous or heterozygous form, the summer squash will produce white fruit regardless of the Y alleles. A cross between white heterozygotes for both genes (WwYy × WwYy) would produce offspring with a phenotypic ratio of 12 white:3 yellow:1 green.
Finally, epistasis can be reciprocal such that either gene, when present in the dominant (or recessive) form, expresses the same phenotype. In the shepherd’s purse plant (Capsella bursa-pastoris), the characteristic of seed shape is controlled by two genes in a dominant epistatic relationship. When the genes A and B are both homozygous recessive (aabb), the seeds are ovoid. If the dominant allele for either of these genes is present, the result is triangular seeds. That is, every possible genotype other than aabb results in triangular seeds, and a cross between heterozygotes for both genes (AaBb x AaBb) would yield offspring with a phenotypic ratio of 15 triangular:1 ovoid.
As you work through genetics problems, keep in mind that any single characteristic that results in a phenotypic ratio that totals 16 is typical of a two-gene interaction. Recall the phenotypic inheritance pattern for Mendel’s dihybrid cross, which considered two non-interacting genes—9:3:3:1. Similarly, we would expect interacting gene pairs to also exhibit ratios expressed as 16 parts. Note that we are assuming the interacting genes are not linked; they are still assorting independently into gametes.
 For an excellent review of Mendel’s experiments and to perform your own crosses and identify patterns of inheritance, visit the Mendel’s Peas web lab.
For an excellent review of Mendel’s experiments and to perform your own crosses and identify patterns of inheritance, visit the Mendel’s Peas web lab.
Mendel postulated that genes (characteristics) are inherited as pairs of alleles (traits) that behave in a dominant and recessive pattern. Alleles segregate into gametes such that each gamete is equally likely to receive either one of the two alleles present in a diploid individual. In addition, genes are assorted into gametes independently of one another. That is, alleles are generally not more likely to segregate into a gamete with a particular allele of another gene. A dihybrid cross demonstrates independent assortment when the genes in question are on different chromosomes or distant from each other on the same chromosome. For crosses involving more than two genes, use the forked line or probability methods to predict offspring genotypes and phenotypes rather than a Punnett square.
Although chromosomes sort independently into gametes during meiosis, Mendel’s law of independent assortment refers to genes, not chromosomes, and a single chromosome may carry more than 1,000 genes. When genes are located in close proximity on the same chromosome, their alleles tend to be inherited together. This results in offspring ratios that violate Mendel's law of independent assortment. However, recombination serves to exchange genetic material on homologous chromosomes such that maternal and paternal alleles may be recombined on the same chromosome. This is why alleles on a given chromosome are not always inherited together. Recombination is a random event occurring anywhere on a chromosome. Therefore, genes that are far apart on the same chromosome are likely to still assort independently because of recombination events that occurred in the intervening chromosomal space.
Whether or not they are sorting independently, genes may interact at the level of gene products such that the expression of an allele for one gene masks or modifies the expression of an allele for a different gene. This is called epistasis.
[link] In pea plants, purple flowers (P) are dominant to white flowers (p) and yellow peas (Y) are dominant to green peas (y). What are the possible genotypes and phenotypes for a cross between PpYY and ppYy pea plants? How many squares do you need to do a Punnett square analysis of this cross?
[link] The possible genotypes are PpYY, PpYy, ppYY, and ppYy. The former two genotypes would result in plants with purple flowers and yellow peas, while the latter two genotypes would result in plants with white flowers with yellow peas, for a 1:1 ratio of each phenotype. You only need a 2 × 2 Punnett square (four squares total) to do this analysis because two of the alleles are homozygous.
Assuming no gene linkage, in a dihybrid cross of AABB x aabb with AaBb F1 heterozygotes, what is the ratio of the F1 gametes (AB, aB, Ab, ab) that will give rise to the F2 offspring?
A
The forked line and probability methods make use of what probability rule?
B
How many different offspring genotypes are expected in a trihybrid cross between parents heterozygous for all three traits when the traits behave in a dominant and recessive pattern? How many phenotypes?
D
Use the probability method to calculate the genotypes and genotypic proportions of a cross between AABBCc and Aabbcc parents.
Considering each gene separately, the cross at A will produce offspring of which half are AA and half are Aa; B will produce all Bb; C will produce half Cc and half cc. Proportions then are (1/2) × (1) × (1/2), or 1/4 AABbCc; continuing for the other possibilities yields 1/4 AABbcc, 1/4 AaBbCc, and 1/4 AaBbcc. The proportions therefore are 1:1:1:1.
Explain epistatis in terms of its Greek-language roots “standing upon.”
Epistasis describes an antagonistic interaction between genes wherein one gene masks or interferes with the expression of another. The gene that is interfering is referred to as epistatic, as if it is “standing upon” the other (hypostatic) gene to block its expression.
In Section 12.3, “Laws of Inheritance,” an example of epistasis was given for the summer squash. Cross white WwYy heterozygotes to prove the phenotypic ratio of 12 white:3 yellow:1 green that was given in the text.
The cross can be represented as a 4 × 4 Punnett square, with the following gametes for each parent: WY, Wy, wY, and wy. For all 12 of the offspring that express a dominant W gene, the offspring will be white. The three offspring that are homozygous recessive for w but express a dominant Y gene will be yellow. The remaining wwyy offspring will be green.

You can also download for free at http://cnx.org/contents/185cbf87-c72e-48f5-b51e-f14f21b5eabd@11.2
Attribution: